1 Introduction
Ancient DNA (aDNA) offers Bioarchaeologists fresh opportunities to tackle some of the biggest archaeological debates to date. But with all the excitement about what this field of study could reveal, it is easy to overlook some core problems in using aDNA. This essay will therefore critically assess the contribution and potential of aDNA analysis to increase our understanding of both the origins and migrations of Homo sapiens, and discuss how aDNA does not necessarily provide the answers.
1.1 Origins
Questions about the origin of our species have been much debated, and in particularly regarding whether humans originate “Out of Africa” or if a “Multiregional Theory” is more likely. aDNA has been able to contribute to this debate through its ability to identify Mitochondrial Eve, as well as showing that the genetic history of Homo sapiens involves a mixture of other hominins. However the potential of aDNA for understanding the origin of Homo sapiens will always be limited. This is due to the reliance on: sex chromosomes to create genetic lineages, genetic sexing, phylogenetic trees, using molecular clocks, and most importantly the preservation of genetic data.
1.2 Migration
Migration is another fascinating phenomenon in which aDNA has attempted to gain an insight. This can be seen in the neolithization of Europe, through evidence of Lactose Persistency, and animal genetics (Itan et al. 2009, 7; Karimi et al. 2016). However, as the research examines the DNA of individuals, thus recognising the high sequencing costs and the destruction of samples due to the invasive process, it may not be representative of society as a whole. The genotyping of animals also cannot necessarily guarantee there has been a migration of people.
2 Origins
2.1 Why use aDNA?
2.1.1 mtDNA and Y chromosome
The process of evolution takes millions of years, with offspring being nearly identical to parents (Darwin 1859, Dawkins 1989). It can be argued that the first human was not human at all but an ape. It has also been argued that the first human was similar to a fish, or a single-celled organism (Dawkins 2012, 50). One method used to understand these origins is aDNA. Genetic lines such as mitochondrial DNA (mtDNA) (inherited maternally), and Y chromosomes (inherited paternally), enable an understanding of the location from which the first humans originate. However, significantly, the mitochondria in both offspring sexes will only be inherited from the mother.
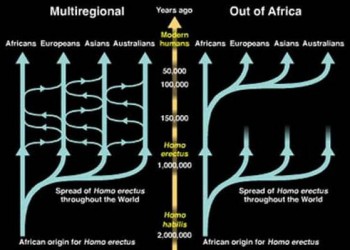
There are two conflicting hypotheses on the origins of Homo sapiens: the “Out of Africa” hypothesis and the “Multiregional Theory”. The first suggests that Homo sapiens originated principally in one location, Africa, before migrating throughout the globe, whilst the latter suggests that there was a level of gene flow in different locations from Homo erectus, as shown by figure 1 (Satta and Takahata 2002, 872). To be able to answer this question, through analysis of the genetic lines, a most recent common ancestor must be established. If the ancestor appears to have originated in Africa, this would provide evidence for the “Out of Africa” hypothesis. Finding this ancestor requires the tracing of all modern DNA to “Mitochondrial Eve” and “Y Chromosome Adam”, and this is the approach upon which many studies have been based (Ke et al. 2001; Pakendorf and Stoneking 2005). The origins of both Mitochondrial Eve and Adam are estimated to be out of Africa, as the genetics are more divergent in Africa than elsewhere, supporting the claim of “Out of Africa” (Vigilant et al. 1991, 253). It is clear that the contribution of aDNA is that not only is it able to identify the species of Homo sapiens, but also provide answers as to where it originates from. Thus, using aDNA shows that Homo sapiens originate from Africa.
2.1.2 Admixture
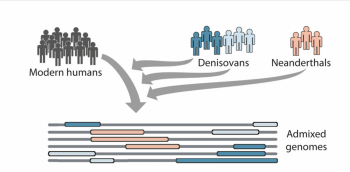
aDNA has also contributed to the debates on the origins of humans, as it has been able to give us more of an understanding of which species Homo sapiens comes from. Through the analysis of the aDNA of anatomically modern humans, Neanderthals and Denisovans, a level of admixture was discovered, meaning that there are genes in modern humans, which originated from these archaic hominins, as shown by figure 2 (Browning et al. 2018, 53). To truly understand the origins of humans, it is necessary to consider Homo sapiens, Neanderthals and Denisovans as being the genetic origins. As a result, It is clear that aDNA has truly revolutionised bioarchaeological understanding of humans as a species. Consequently, aDNA has contributed to the question of origin, as it has been able to show that the genetic history of modern humans involves other Hominin species.
2.2 The issues
2.2.1 Mitochondria
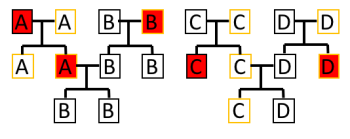
Opinions on the reliability of Mitochondrial Eve have been fluctuating since its first conception (Gibbons 1992, 873). There are many reasons for this, however, this essay will focus on the misunderstanding of Mitochondrial Eve and the recent discovery of male line mitochondria inheritance. Firstly, being able to identify the origin of Mitochondrial Eve does not mean the identification of the first human (Learn, 2016). As shown by figure 3, sex of offspring and genus origin yields a complex and extensive genetic lineage that ultimately obscures the culminated admixture. This is due to the individual lineages being inherited from a particular sex, so if the offspring are of the same sex, or there is no offspring, the genetic lineages will stop. As a result, this study does not lead to the first human; rather it leads to the survival of one genetic line through gene flow (ibid). Consequently, it is apparent that arguments using Mitochondrial Eve to locate the origin of Homo sapiens are inaccurate, and thus the potential of aDNA to understand the origin is simply too unreliable. Secondly, recent studies have revealed that Mitochondria can also be inherited by the father (Luo et al. 2018, 13044). As this research has been published after much research into Mitochondrial Eve, it paints uncertainty thus showing that aDNA may not have such a great potential as first thought.
2.2.2 Y chromosome
2.2.2.1 Biological sexing
There are similarly a range of issues associated with the Y chromosome. The first of these is the identification of genetic sex. To be able to work out the sex chromosome to test, identification of the genetic sex of the individual is needed. Seeing as the Y chromosome is only present in males, by identifying the presence or the difference between AMELX and AMELY can determine the sex. These are relatively inaccurate, however, as the absence of Y chromosome can be caused by a female sample, DNA degradation or an inhibitor in the sample. Amelogenin also has its issues in that allelic dropout, in which the longer chain, i.e. the Y chromosome, is not detectable using PCR techniques. Further issues arise from chromosomal anomalies in which the individual may have phenotypic differences to the chromosomal presence (Brown and Brown 2011, 158-161). A final difficulty in using genetic sex is that despite genetically it may be typed for a particular sex, phenotypically gender cannot be determined entirely. Henceforth, it may be difficult to infer the life that the individual had (Short et al. 2016, S93).
2.2.2.2 Ambiguity
Issues with the inability to identify the individual’s sex, creates issues due to the ambiguity of which genetic line is to be tested. Tests themselves are expensive and this may cause possible samples to be discarded as they cannot be certain of the sex. Although the mitochondria will be inherited in both males and females from the mother, the Y chromosome will not be typed due to the difficulty in determining its presence as well as the fragility of the long chains. These elements should certainly be reconsidered before conducting future investigations into aDNA, lest we continue to provide data with inaccuracies and increase misrepresentative interpretations of individuals regarding sex demographic versus potential complex gender social constructs
2.2.2.3 Mutations
Mutations also play their role. Mutations mainly happen in the production of gametes for sexual reproduction. The rate of mutation is higher for male gametes due to the rapid transcription and translation of the parent DNA strand to the male gamete from one initial strand thousands of times, increasing the chance of mutation (Wilson Sayres and Makova 2011, 939). It is therefore hypothetically possible that the mutations will mean that the identification of genetic lineages using Y chromosome is more open to inaccuracies due to this increase in mutations.
2.2.3 Phylogenetic trees
A final issue in finding the origin of Homo sapiens is the use of phylogenetic trees. This process works by investigating the diversity of different samples based on different genetic morphometrics. Molecular clocks are needed for this study to take place. Several controls must be put in place, such as consistent demography, mutation rate and reproduction (Moorjani et al. 2016, 10607–10608). These controls are not reliable in reality. This is because demographics change throughout time and, as there is no accurate measure to estimate population, the rate of mutation relies on generalisations which oposes against the nature of mutations. Also, reproductive ages have been changing throughout time, particularly in the last 100 years, within the time in which DNA sequencing has taken place (Hassan 1978, 49; Lynch 2010, 345; Toulemon 1988, 6). The reliance on controls to be able to use molecular clocks shows that aDNA in conjunction with phylogenetic trees is simply not viable, accordingly the potential and contribution of these studies is limited.
2.2.4 Preservation
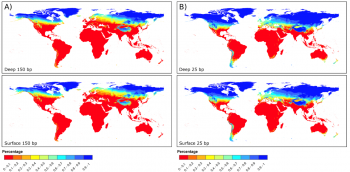
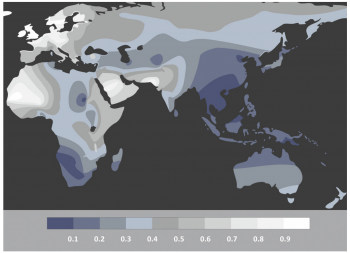
However, the above discussion relies on the discovery of samples. As is often said in archaeology; absence of evidence is not evidence of absence. Seeing as the current understanding of “Out of Africa” implies the origins coming from an area of hot climates, the chances of organic remains containing aDNA preservation is less than 0.1 in Africa, as shown by figure 4. Accordingly, it could be possible that the full understanding of the origins of Homo sapiens will never be fully understood. An example of this is Adcock et al. (2001 538), who proposed that a different mtDNA group was the most divergent, based on the findings of Lake Mungo. This claim was later dismissed by Heupink et al. (2016, 6893) who stated there was no Aboriginal Australian DNA but rather it came from modern European contamination, disproving this research. This research demonstrates that there may now be samples which would revolutionise the understanding of the origins of Homo sapiens. When combined with new discoveries such as Jebel Irhoud, Morocco, which pushed the date of Homo sapiens’ origin further back, it poses the possibility that there are human remains that are yet to be discovered (Richter et al. 2017, 296). Due to the lack of preservation, the contribution of aDNA may never give a full understanding as to the origin of Homo sapiens. With oxidation preventing the full transcription of genetic coding combined with the scarcity of samples fit for testing, it becomes explicit that human origin will never be fully understood.
3 Migrations
3.1 Case study: Lactase persistence
Since the origin of Homo sapiens, migrations have enabled humans to occupy six of the seven continents. These have happened in a range of significant events. Figure 5 shows the current understanding of how these spreads took place. To be able to show the limitations of aDNA research in relation to migration case studies of lactase persistence, the populating of America, and the process of Neolithization will be used to illustrate the points.
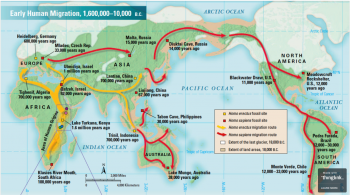
3.2 Why use aDNA?
Lactase persistence is a haplogroup that has arisen in five different ways, reaching 35% of people. In Europe, it has been proposed to have arisen around 6256 to 8683 BP in North Balkans and Central Europe, probably at the same time as the development of dairying (Itan et al. 2009, 7). Figure 6 shows the percentage of people with lactase persistence in different locations. As this is a genetic trait, it is possible to deduce the spread, thus migrations, of people using aDNA techniques. A paper by Krüttli et al. (2014 e86251) suggests that by Medieval times, the spread of lactase persistence (LP) had encompassed the whole of Europe. This was discovered by sequencing a range of samples, of which 70% of the sample was LP (2014, 3). This paper shows the potential of aDNA for archaeological questions it is able to show the level of migrations of people and the interactions of different groups in a way which is inaccessible by different methods. Consequently, aDNA has the potential to contribute to the understandings of migrations.
3.3 The issues
3.3.1 Individuality
To be able to evaluate the migrations, aDNA analysis studies rely on individuals. As a result, the first question is, are these individuals representative of society? If these individuals are anomalous to the society at the time, the results will present a different analogy to the actual trend. In regards to LP, this can be shown in a paper by Malmström et al. (2010, 3), which used four different sites all located on one island off the coast of Sweden, with ten results, to show that the modern distribution of LP in Scandinavia is not due to the haplogroup originating in this area. Seeing as LP is a single polymorphism, which is dominant and can be caused by a mutation, it is theoretically possible that these results were caused by other factors rather than the spread of LP to this region, for example, it may be a regional anomaly (Swallow 2003, 197). The uncertainty as to whether the sample is representative of a society is made more of an issue due to the high costs and destructive nature of aDNA analysis. As a result, it is not feasible to carry out analysis on more samples. Through these examples of individuality, the overall contribution potential of aDNA to understand migrations is therefore limited by accessibility to analyse more results, to counteract the individuality of results.
3.3.2 Trading domestication
By using the DNA changes between domesticated and wild populations, it is proposed that the spread of neolithization across Europe will be further understood. Many studies have been based on understanding the genetic changes between local wild and introduced domesticated animals and plants, trying to understand the interactions and the level of admixture (e.g. Karimi et al. 2016). Through studying the domesticated relationships, this can be argued to represent the movement of people throughout Europe at this time (Larson and Burger 2013, 198). This should, however, not be claimed as the case. The movement of animals and varieties of crops do not necessarily mean the movement of people. This is as “the spread of agriculture involved a variety of mechanisms and cannot be merely explained by a simple model of migration or acculturation” (Divišováa 2012, 149). To give an example, it may be possible that livestock or products of crops were traded between neighbouring groups, which would argue the spread of the Neolithic “package” instead of people. This clearly points out that following the genetics of animals alone cannot guarantee a reliable method of tracking the migration of people themselves.
4 Analysis
aDNA analysis has developed a lot in the last few years in terms of its capabilities and use for ground-breaking discoveries. There are, however, many complications which arise which renders the capability and potential of such research techniques not as clear cut and decisive.
4.1 Origins
aDNA has contributed to the debates on origins by first defining what being human is as well as discovering Homo sapiens are part Neanderthal and Denisovan. Despite this contribution, factors such as misunderstandings of Mitochondrial Eve not being the first human, mitochondria being inherited paternally, difficulties in sexing, use of controls for molecular clocks which are factually inaccurate, and preservation of genetic material all mean that the potential for using aDNA to understand the origins of Homo sapiens will always be limited. Consequently, it could be argued that aDNA can never reliably contribute to the understanding of Homo sapiens origins.
4.2 Migration
aDNA can contribute to the debates of migrations as not only can aDNA discover information which is inaccessible through alternative methods, but also allows the discoveries to be tested in terms of the reliability of the results. Despite this, contamination, issues of individuality, and the use of animals to understand human relationships, means that studies using aDNA will never escape the clutches of scepticism.
4.3 Conclusion
It is through these case studies that an understanding of the limitations of aDNA to the large debates in archaeology can be accessed, and through time, it will be possible to develop new techniques with new genetic material to understand the lives of our ancestors.





![Figure 1: Map of cemeteries studied in this essay. Porter, C, "Map of Cemeteries Used" [JPG map], Scale 1:100000, GB Overview [ geospatial data], Updated: 2019, Ordnance Survey (GB), Using: EDINA Digimap Ordnance Survey Service, <http://digimap.edina.ac.uk/>, Created: October 2019.](https://theposthole.org/sites/theposthole.org/files/styles/article_thumb/public/articles/454/fig%202.png?itok=cGHEw2KW)
